Lu Zhang
About Me
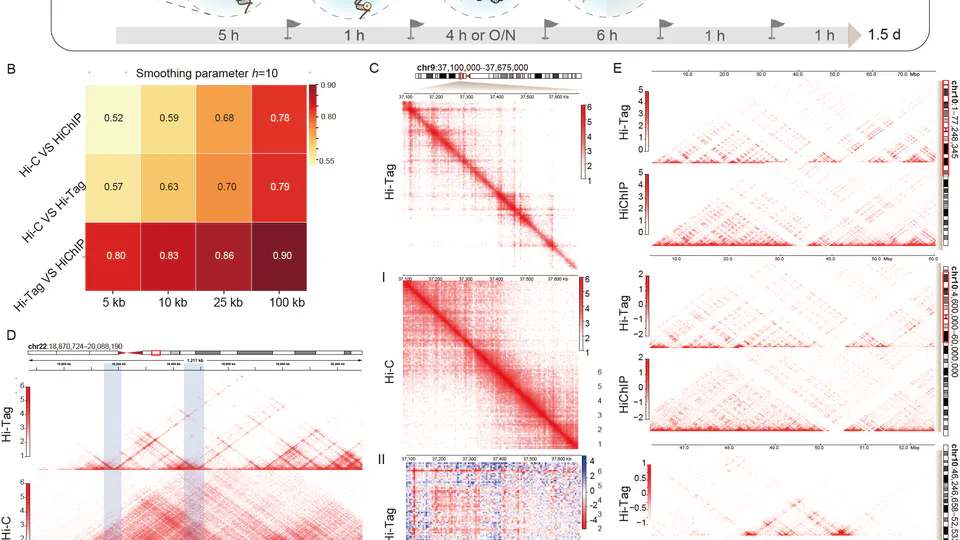
I am a postdoctoral research scholar in Dr.Anna Gloyn’s Translational Genomics of Diabetes Lab. During my master’s and doctoral studies, I focused on epigenomics and single-cell multi-omics analysis, with a particular emphasis on 3D genomics. My research included developing Hi-Tag, a chromatin conformation capture technique designed specifically for small cell samples. This method provides valuable insights into the organization of chromatin within the cell.
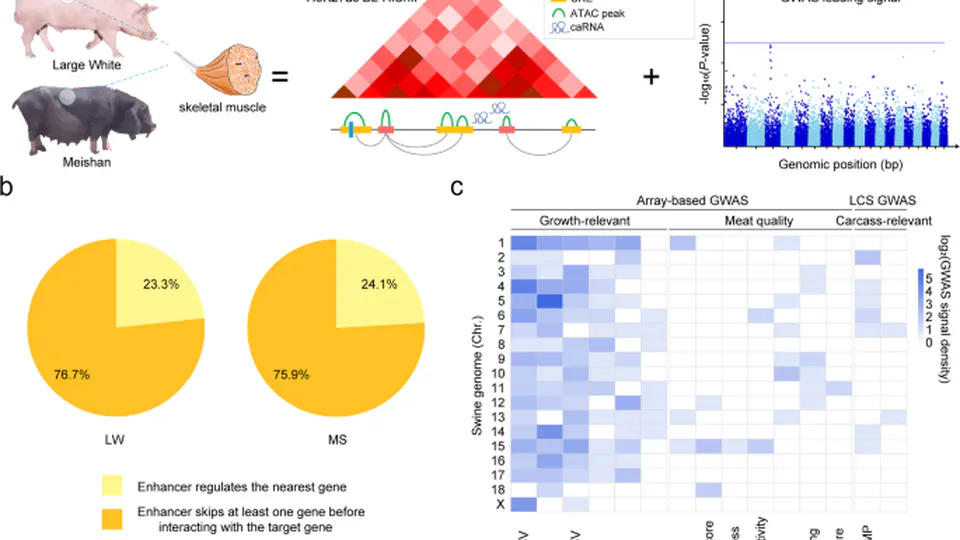
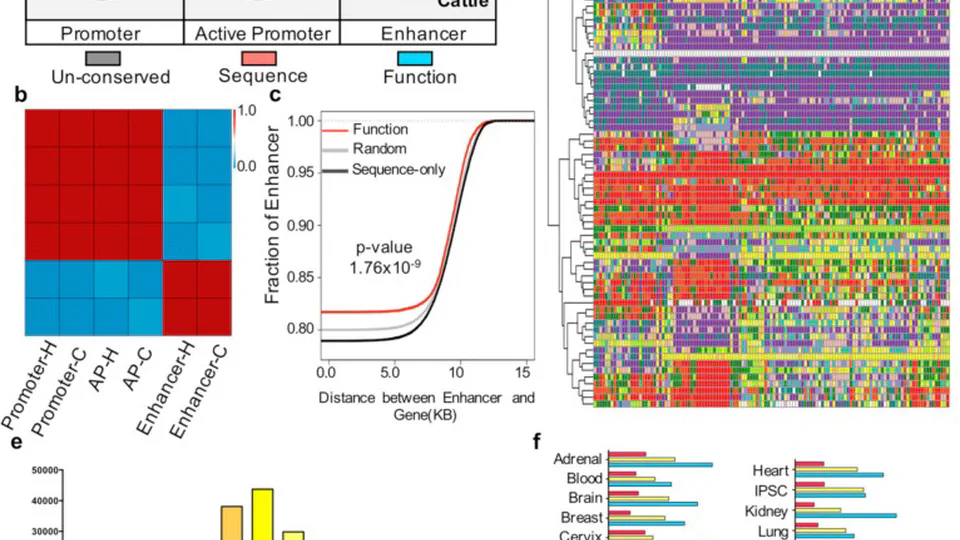
I have built strong expertise in integrating various types of biological data, including RNA-seq, ATAC-seq, chromatin interaction data, and single-cell data. I have also contributed as a co-author to several research projects, including studies that utilized genome-wide association studies (GWAS) and GTEx data to link multi-omics data with functional genomics. These experiences have allowed me to develop a deep understanding of how to integrate diverse genomic data to address complex biological questions.
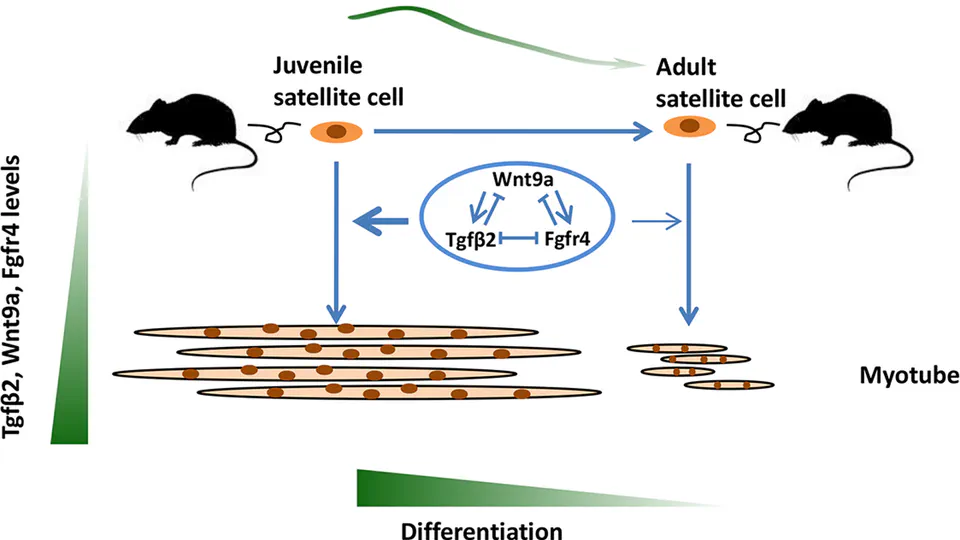
Currently, I am applying my research skills to explore questions related to diabetes.
- Three-dimensional genomics
- Single-cell multi-omics
- Spatial omics
- Deep Learning in Genomics
PhD in Animal Genetics, Breeding, and Reproduction
Huazhong Agricultural University
Visiting PhD in Animal Genetics, Breeding, and Reproduction
Center for Quantitative Genetics and Genomics, Aarhus University
Exchange Student in Animal Science
South China Agricultural University
BSc in Animal Science
ShiHeZi University